Triturus arntzeni
|
publication ID |
https://doi.org/10.11646/zootaxa.3682.3.5 |
|
publication LSID |
lsid:zoobank.org:pub:69B9A846-616F-4774-B3F0-B796D2B90431 |
|
DOI |
https://doi.org/10.5281/zenodo.5658359 |
|
persistent identifier |
https://treatment.plazi.org/id/EB5087CA-FF93-530F-00DB-4EC2F0CCF489 |
|
treatment provided by |
Plazi (2016-04-14 14:15:03, last updated 2024-11-29 17:26:04) |
|
scientific name |
Triturus arntzeni |
| status |
|
The name T. arntzeni is a junior synonym of T. macedonicus
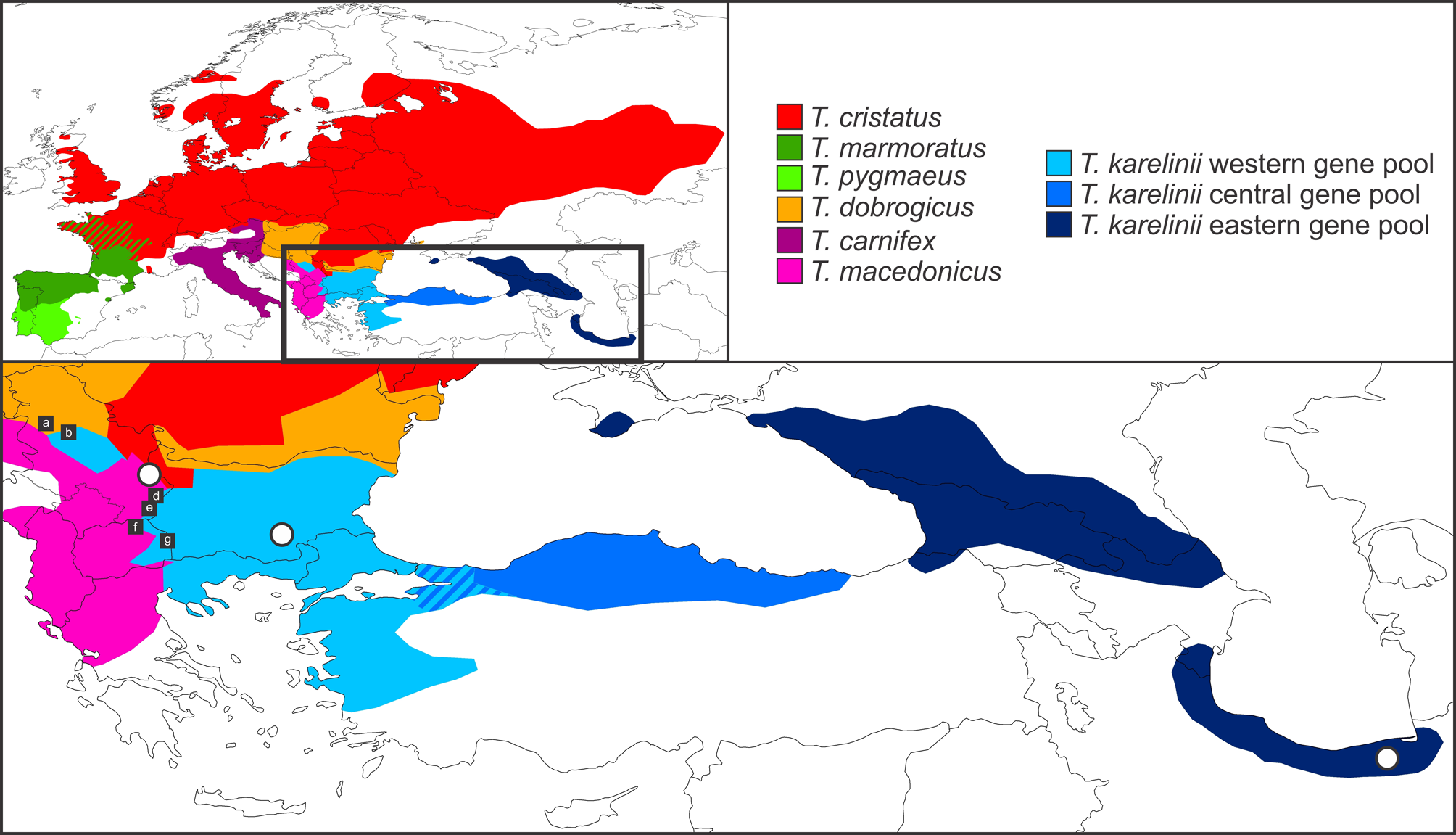
Litvinchuk et al. (1999) described the subspecies Triturus karelinii arntzeni based on differences in genome size, protein variation and morphological characteristics. Subsequently, Espregueira Themudo et al. (2009) elevated this subspecies to species level, i.e. Triturus arntzeni Litvinchuk, Borkin, Džukiċ and Kaleziċ, 1999 (in Litvinchuk et al., 1999). However, concern has been expressed that the type locality of T. arntzeni —Vrtovaċ, near Pirot in eastern Serbia ( Fig. 1 View FIGURE 1 )—in fact represents the species T. macedonicus and not T. karelinii s. l. (Arntzen & Wielstra, 2010; Stoyanov et al., 2011). We review the species identity of crested newts from Vrtovaċ below. Because the bulk of evidence points towards T. macedonicus being the crested newt species occurring at Vrtovaċ, we identify the name T. arntzeni as a junior synonym of T. macedonicus and conclude that it does not represent a taxon newly distinguished in T. karelinii s. l.
Geography. The population in Litvinchuk et al. (1999) representing T. arntzeni comprises seven different localities (14a-g; see Fig. 1 View FIGURE 1 ), all of which are situated at, or close to, various crested newt contact zones. Note that locality 14c concerns the actual type locality of T. arntzeni . For the latest overview of Triturus distribution, including a database of locality data, see Wielstra et al. (2013b). The species composition of the seven T. arntzeni populations we evaluate as follows:
Locality 14a, Trbusaċ, Serbia is T. dobrogicus ( Wielstra et al., 2013b). Species identification is based upon the phenotype of material deposited at the Institute of Biological Research, “Siniša Stankoviċ”, University of Belgrade (JWA, BW).
Locality 14b, Trešnja, Serbia represents the western T. karelinii s. l. nuclear gene pool with T. dobrogicus mitochondrial DNA and some limited presence of, or genetic admixture with, T. dobrogicus (Arntzen et al., submitted; Wallis & Arntzen, 1989).
Locality 14c, Vrtovaċ, Serbia, is positioned inside the T. macedonicus range (Wielstra & Arntzen, 2012). Neighboring crested newt species are, in order of increasing distance, T. cristatus , T. dobrogicus and western T. karelinii s. l. nuclear gene pool ( Fig. 1 View FIGURE 1 ).
Locality 14d, Vlasi, Serbia, has western T. karelinii s. l. nuclear gene pool and T. macedonicus in syntopy (Wielstra & Arntzen, 2012). Species identification is based on examination of animals observed in the field (JWA, BW).
Locality 14e, Vlasina, Serbia has T. macedonicus (Wielstra & Arntzen, 2012) . Species identification is based upon the phenotype of material deposited at the Institute of Biological Research, “Siniša Stankoviċ”, University of Belgrade and of animals observed in the field (JWA, BW). The number of rib-bearing vertebrae (details below) reported in this population (n=7 with NRBV = 13, n=25 with NRBV = 14) is typical for T. macedonicus ( Crnobrnja-Isailoviċ et al., 1997) .
Locality 14f, Stracin, Macedonia has the western T. karelinii s. l. nuclear gene pool and T. macedonicus in syntopy (Wielstra & Arntzen, 2012). Species identification is based upon the phenotype of material deposited at the Institute of Biological Research, “Siniša Stankoviċ”, University of Belgrade (JWA, BW). Note that a population of T. macedonicus is located c. 4 kilometers to the west (Rugince, Macedonia in Wielstra & Arntzen, 2012).
Locality 14g, Berovo, Macedonia is western T. karelinii s. l. nuclear gene pool. Species identification was confirmed based upon the phenotype of material deposited at the Institute of Biological Research, “Siniša Stankoviċ”, University of Belgrade and of animals observed in the field (JWA, BW) and conforms to species identification by Kaleziċ and Hedgecock (1980). Another confirmed T. karelinii s. l. nuclear gene pool locality is located 9 kilometers to the northwest (Mitrašinci, Macedonia in Wielstra et al., 2013a).
Throat and belly pattern. In the field crested newts are best identified by morphotype (see introduction) and by their coloration pattern ( Arntzen, 2003; Arntzen & Wallis, 1999) as follows:
Triturus karelinii s. l.: Stocky build. Heavily white-stippled sides. Ventral surface yellow-orange with many small to medium-sized, frequently angular black spots and continuous on the throat.
Triturus carnifex: Medium stocky build. Little or no white stippling on sides. Throat color variable with white stipples. Ventral surface yellow with few large, roundish, ill-defined and muddy-gray to black spots.
Triturus macedonicus: Medium stocky build. Sides densely white-stippled. Throat dark black or a muddied mix of black and yellow with many, medium sized white stipples. Ventral surface yellow to orange-yellow with a dense pattern of small, irregular spots.
Triturus cristatus: Slender build. Heavily white-stippled sides. Throat a muddied mix of black and yellow with fine white stippling. Ventral surface yellow-orange with irregular black spots.
Triturus dobrogicus: Very slender build. Heavily white-stippled sides. Black throat with large angular white spots (especially in males), ventral surface deep orange with many sharp, roundish black spots.
For a picture overview of throat and belly patterns of the different crested newt species, see Arntzen and Wallis (1999). Pictures of the throat and belly pattern of the holotype and paratypes of T. arntzeni (online Appendix 1), preserved material stored at the Institute of Biological Research, “Siniša Stankoviċ”, University of Belgrade, and live newts from the type locality as observed by several of us (JWA, BW) most resemble T. macedonicus and not T. karelinii s. l. In particular we point to the absence of the distinctive T. karelinii s. l. throat pattern, with small angular dark spots surrounded by yellow, similar to the pattern on the belly. These spots are also absent in the holotype of T. arntzeni . To ascribe the Vrtovaċ newts to T. macedonicus by coloration pattern alone is not straightforward: as noted by Freytag (1988), T. macedonicus overlaps considerably with T. cristatus in patterning and coloration.
Number of rib-bearing pre-sacral vertebrae. The Triturus morphotypes were initially distinguished with the help of the ‘Wolterstorff index’, a measure of overall shape, defined as ‘forelimb length divided by inter-limb distance’ ( Wolterstorff, 1923). Arntzen and Wallis (1994) found the number of rib-bearing pre-sacral vertebrae (NRBV) in Triturus to represent a taxonomically discriminating character superior to the Wolterstorff-Index (the two are strongly negatively correlated across species). Accordingly, the Triturus morphotypes are characterized by a modal number of rib-bearing pre-sacral vertebrae (NRBV) ( Arntzen & Wallis, 1999). Modal NRBV count increases from 12 in T. marmoratus – T. pygmaeus , via 13 in T. karelinii s. l., 14 in T. carnifex – T. macedonicus and 15 in T. cristatus , to 16/ 17 in T. dobrogicus ( Arntzen, 2003; Arntzen & Wallis, 1999).
A disadvantage of NRBV (similarly applying to the Wolterstorff index) is that, although informative at the population level, it is not unambiguous at the individual level, due to intraspecific variation. Moreover, intermediate scores due to interspecific hybridization may point to a third species not involved ( Arntzen & Wallis, 1994). We determined NRBV for 63 individuals from the T. arntzeni type locality (the holotype and 12 paratypes, deposited at the Zoological Institute, Russian Academy of Sciences, St. Petersburg; 46 specimens deposited at the Institute of Biological Research, “Siniša Stankoviċ”, University of Belgrade and four specimens deposited at Naturalis Biodiversity Center, Leiden, see online Appendix 2). Three specimens with different NRBV (13 versus 14) on either sides of the body were ignored. Eleven individuals (18.3%) have an NRBV count of 13, NRBV is 14 in n=44 (73.3%) and NRBV is 15 in n=5 (8.3%). The distribution of modal versus non-modal NRBV scores in the Vrtovaċ population is not significantly different from that of T. carnifex / T. macedonicus (G-test of independence, d. f. = 1, P>0.05; reference data in Arntzen, 2003). The NRBV count of 15 shown by the holotype of T. arntzeni is indicative since this value fits T. macedonicus better than it does T. karelinii s. l.
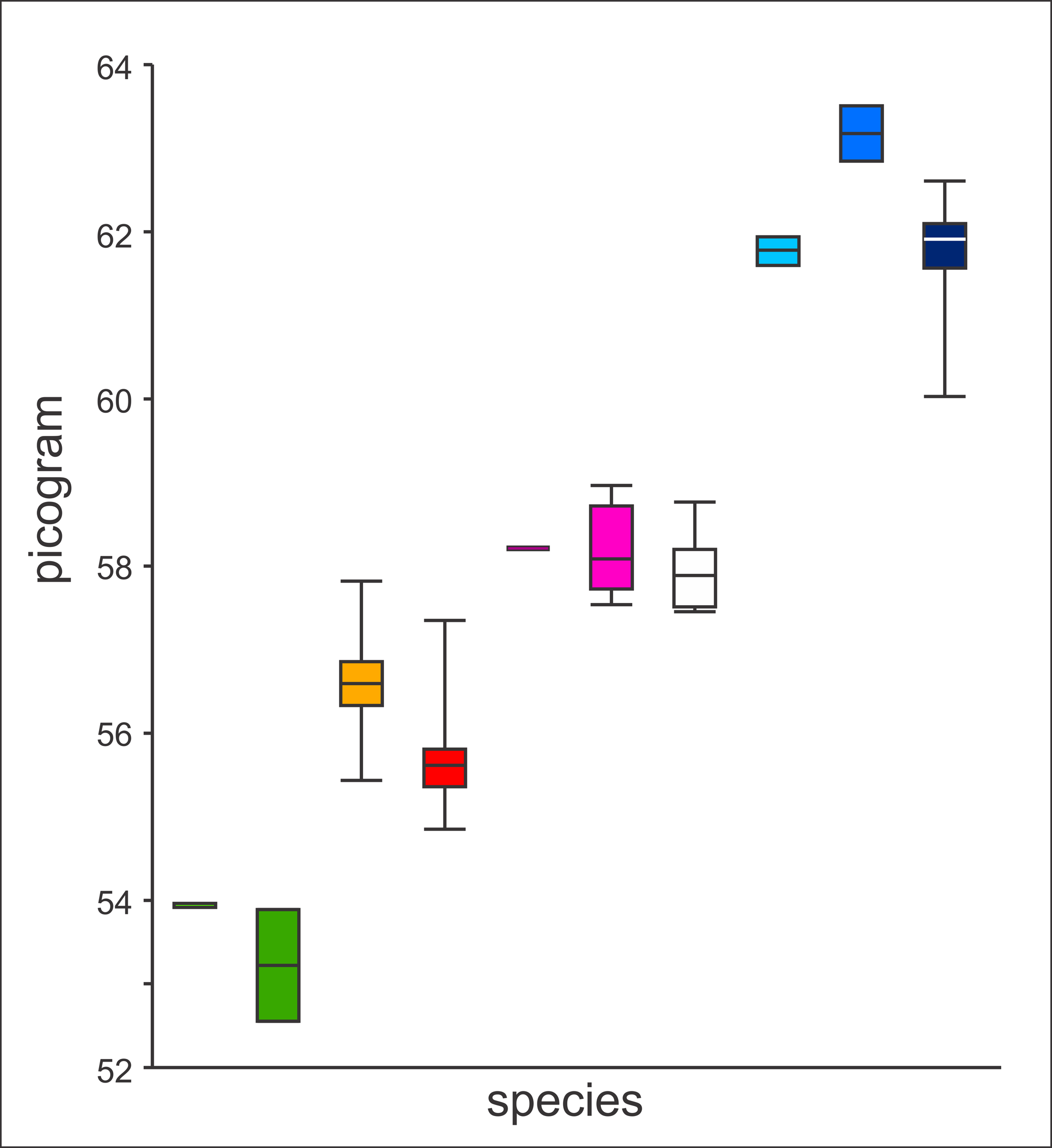
Genome size. The size of the genome may be a source of relevant taxonomic information ( Green & Sessions, 2007; Kron et al., 2007). Litvinchuk et al. (1999) observed a difference in genome size between T. arntzeni and T. karelinii s. l., with non-overlapping confidence intervals. We here interpret the genome size of T. arntzeni from Vrtovaċ (n=13) not as ‘different from other T. karelinii’ (as in Litvinchuk et al., 1999), but as ‘not different from T. macedonicus’ ( Fig. 2 View FIGURE 2. A ; constituent data in online Appendix 3). It is important to note that these genome sizes also differ from those of T. cristatus and T. dobrogicus . In contrast to Litvinchuk et al. (1999) we do not invoke convergent evolution or introgressive hybridization to explain the data but conclude that newts from Vrtovaċ actually are T. macedonicus . The genome size of the holotype of T. arntzeni in particular falls within the range of T. macedonicus but outside of the range of T. karelinii s. l. The fourteenth specimen labeled as T. arntzeni in Litvinchuk et al. (1999) is from Tresnja (population 14b, see above) and has a genome size that is intermediate of T. karelinii s. l. and T. dobrogicus ; it is excluded from Fig. 2 View FIGURE 2. A .
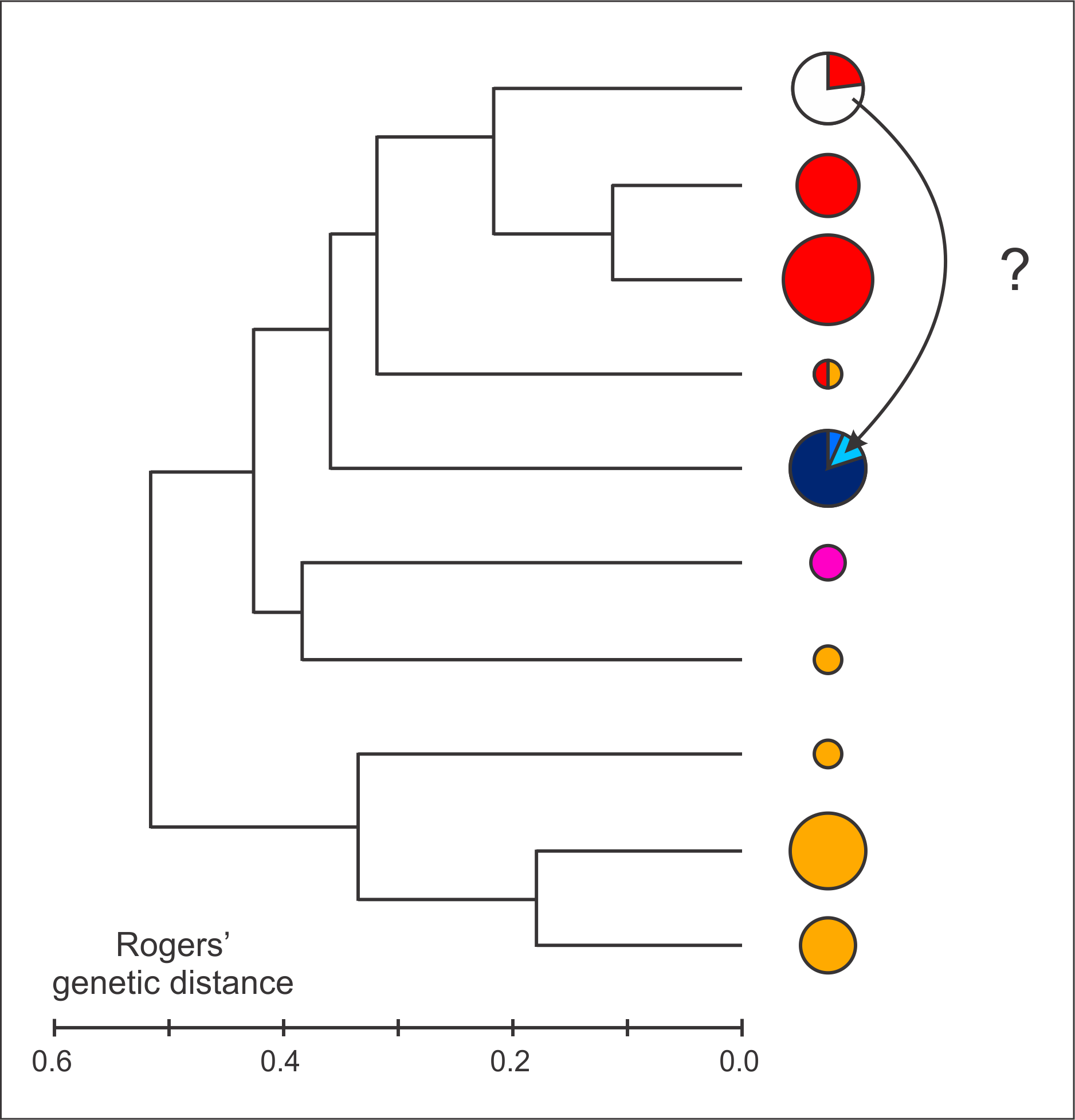
Allozyme data. Litvinchuk et al. (1999) presented data for nine allozyme loci and analyzed these at the population level. We here re-analyze the data at the level of the individual and in a phylogenetic framework for 42 individuals with a complete dataset and 48 individuals with data missing at one locus (online Appendix 4). Unfortunately the dataset for the holotype of T. arntzeni is highly incomplete, but several paratypes can be included in the analysis. We first determine the number of distinctive gene pools that is best supported by the data probabilistically using BAPS v.5.3 ( Corander et al., 2008). The recognized groups were clustered in a dendrogram on the basis of Rogers’ genetic distance with BIOSYS-1 ( Swofford & Selander, 1981).
All ten included T. arntzeni individuals and three T. cristatus are in one group. Other BAPS groups are composed as follows: all T. karelinii s. l. individuals with representatives of all three T. karelinii s. l. nuclear gene pools, the n=3 included T. macedonicus individuals, two T. cristatus groups and four T. dobrogicus groups and, finally, a mixed group with one T. cristatus and one T. dobrogicus (online Appendix 4). In the UPGMA dendrogram ( Fig. 3 View FIGURE 3 ) the BAPS group that contains all T. arntzeni clusters with T. cristatus and it does not cluster with either T. karelinii s. l. or T. macedonicus . A distance-Wagner tree yields the same result (data not shown). We further note that the two loci considered to distinguish T. arntzeni from T. karelinii s. l. by Litvinchuk et al. (1999) are actually represented by the same alleles in T. cristatus and T. macedonicus . However, the reference sample size is small, in particular for unquestionable T. macedonicus (n=3) and western T. karelinii s. l. nuclear gene pool Nuclear DNA sequence data. Wielstra et al. (2013a) provided a framework to cluster Triturus newts to species based on three nuclear DNA markers developed by Espregueira Themudo et al. (2009). Unfortunately, our attempts to obtain nuclear DNA sequence data from the type material (the holotype and 12 paratypes) of T. arntzeni were not successful, presumably because of degraded DNA. However, Wielstra et al. (2013a) presented sequence data for another five individuals from Vrtovaċ (voucher ID 2533-37 stored at Naturalis Biodiversity Center, Leiden). These individuals were all assigned to T. macedonicus (see Appendix S 1 in Wielstra et al., 2013a).
Mitochondrial DNA. As noted above we did not manage to obtain usable DNA extract and were not able to sequence mitochondrial DNA for the type material. However, we note that mitochondrial DNA could not be used to identify these newts anyway. This is because T. macedonicus in and around Serbia contain asymmetrically introgressed western T. karelinii s. l. mitochondrial DNA (the area for which this phenomenon has been documented is detailed in Wielstra & Arntzen, 2012). Similarly, the nearest Serbian T. cristatus populations contain asymmetrically introgressed western T. karelinii s. l. mitochondrial DNA (Wielstra & Arntzen, 2012; Wielstra et al., 2013b). So, from the perspective of mitochondrial DNA, T. macedonicus and T. cristatus are indistinguishable in the Vrtovaċ region.
In conclusion, our analyses confirm that newts from Vrtovaċ, the type locality of T. arntzeni , including the holotype of T. arntzeni , are dissimilar to T. karelinii s. l. and resemble T. macedonicus (NRBV, genome size, nuclear DNA sequences) or T. cristatus (allozyme data). The throat and belly pattern suggest they are T. macedonicus , but we acknowledge that the distinction with T. cristatus is difficult. On balance, we consider the population from Vrtovaċ to represent T. macedonicus (with perhaps some influences of T. cristatus ). Consequently, we consider the name T. arntzeni to be a junior synonym of T. macedonicus and we conclude that the name T. arntzeni should not be used for the central/western T. karelinii s. l. species.
Litvinchuk, S. N., Borkin, L. J., Dzukic, G., Kalezic, M. L., Khalturin, M. D. & Rosanov, J. M. (1999) Taxonomic status of Triturus karelinii on the Balkans, with some comments about other crested newt taxa. Russian Journal of Herpetology, 6, 153 - 163.
Espregueira Themudo, G., Wielstra, B. & Arntzen, J. W. (2009) Multiple nuclear and mitochondrial genes resolve the branching order of a rapid radiation of crested newts (Triturus, Salamandridae). Molecular Phylogenetics and Evolution, 52, 321 - 328. http: // dx. doi. org / 10.1016 / j. ympev. 2009.03.024
Wielstra, B., Crnobrnja-Isailovic, J., Litvinchuk, S. N., Reijnen, B. T., Skidmore, A. K., Sotiropoulis, K., et al. (2013 b) Tracing glacial refugia of Triturus newts based on mitochondrial DNA phylogeography and species distribution modeling. Frontiers in Zoology, 10, 13. http: // dx. doi. org / 10.1186 / 1742 - 9994 - 10 - 13
Wielstra, B., Baird, A. B. & Arntzen, J. W. (2013 a) A multimarker phylogeography of crested newts (Triturus cristatus superspecies) reveals cryptic species. Molecular Phylogenetics and Evolution, 67, 167 - 175. http: // dx. doi. org / 10.1016 / j. ympev. 2013.01.009
Arntzen, J. W. & Wallis, G. P. (1994) The ' Wolterstorff Index'and its value to the taxonomy of the Crested Newt superspecies. Abhandlungen und Berichte fur Naturkunde, 17, 57 - 66.
Arntzen, J. W. & Wallis, G. P. (1999) Geographic variation and taxonomy of crested newts (Triturus cristatus superspecies): Morphological and mitochondrial data. Contributions to Zoology, 68, 181 - 203.
Arntzen, J. W. (2003) Triturus cristatus Superspecies - Kammolch-Artenkreis (Triturus cristatus (Laurenti, 1768) - Nordlicher Kammolch, Triturus carnifex (Laurenti, 1768) - Italienischer Kammolch, Triturus dobrogicus (Kiritzescu, 1903) - Donau- Kammolch, Triturus karelinii (Strauch, 1870) - Sudlicher Kammolch). In: Grossenbacher, K. & Thiesmeier, B. (Eds.), Handbuch der Reptilien und Amphibien Europas. Schwanzlurche IIA. Aula-Verlag, Wiebelsheim, pp. 421 - 514.
Corander, J., Marttinen, P., Siren, J. & Tang, J. (2008) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics, 9, 539. http: // dx. doi. org / 10.1186 / 1471 - 2105 - 9 - 539
Crnobrnja-Isailovic, J., Dzukic, G., Krstic, N. & Kalezic, M. L. (1997) Evolutionary and paleogeographical effects on the distribution of the Triturus cristatus superspecies in the central Balkans. Amphibia-Reptilia, 18, 321 - 332. http: // dx. doi. org / 10.1163 / 156853897 x 00378
Freytag, G. E. (1988) Erinnerungen an Willy Wolterstorff. Wie kompliziert ist die Rassengliederung des Kammolches (Triturus cristatus [LAURENTI, 1768])? (Amphibia, Caudata, Salamandridae). Ein blick in die Geschichte der Salamanderkunde. Zoologische Abhandlungen fur Tierkunde Dresden, 44, 1 - 10.
Green, D. M. & Sessions, S. K. (2007) Chapter 6 - Karyology and cytogenetics. In: Heatwole, H. (Ed), Amphibian Biology Volume 7. Surrey Beatty & Sons, Chipping Norton, NSW, Australia, pp. 2757 - 2842.
Kalezic, M. L. & Hedgecock, D. (1980) Genetic variation and differentiation of three common European newts (Triturus) in Yugoslavia. British Journal of Herpetology, 6, 49 - 57.
Kron, P., Suda, J. & Husband, B. C. (2007) Applications of flow cytometry to evolutionary and population biology. Annual Review of Ecology Evolution and Systematics, 38, 847 - 876. http: // dx. doi. org / 10.1146 / annurev. ecolsys. 38.091206.095504
Stoyanov, A., Tzankov, N. & Naumov, B. (2011) Die Amphiben und Reptilien Bulgariens. Chimaira, Frankfurt am Main, 582 pp.
Swofford, D. L. & Selander, R. B. (1981) BIOSYS- 1: A FORTRAN program for the comprehensive analysis of electrophoretic data in population genetics and systematics. Journal of Heredity, 72, 281 - 283.
Wallis, G. P. & Arntzen, J. W. (1989) Mitochondrial-DNA variation in the crested newt superspecies: Limited cytoplasmic gene flow among species. Evolution, 43, 88 - 104. http: // dx. doi. org / 10.2307 / 2409166
Wolterstorff, W. (1923) Ubersicht den Unterarten und Formen des Triton cristatus Laur. Blatter fur Aquarien-und Terrarienkunde, 34, 120 - 126.
FIGURE 1. The distribution of Triturus karelinii s. l. The inset shows the distribution of Triturus (the red / green interdigitating shades refer to an area where T. marmoratus and T. cristatus are distributed sympatrically) and the cut-out shows the distribution of the three nuclear gene pools comprising T. karelinii s. l. in detail. The interdigitating shades of blue represent the area in which the central T. karelinii s. l. nuclear gene pool carries western T. karelinii s. l. mitochondrial DNA. The three white dots in the cut-out show type localities with, from left to right, Vrtovać, Serbia for T. arntzeni, Ostar Kamak, Bulgaria for T. ivanbureschi sp. nov. and the coast of the Gulf of Gorgan, Iran for T. karelinii sensu stricto. The black squares show six of the seven populations (a-b and d-g) representing locality 14 of T. arntzeni in Litvinchuk et al. (1999 )); the left white dot corresponds to the seventh locality (c; see text for details).
FIGURE 3. Genetic similarity among crested newts (Triturus cristatus superspecies) based on the allozyme data presented in Litvinchuk et al. (1999). Similarity among ten groups of crested newt, identified by Bayesian clustering, is expressed in an UPGMA dendrogram built with Rogers’genetic distance (for details see text and for data see online Appendix 4). Pie sizes reflect sample size. Color codes are as in Fig. 1 and individuals from the population of special interest (the type locality of T. arntzeni — Vrtovać) are shown in white. Note that this group does not cluster with T. karelinii s. l.
FIGURE 2. A comparison of the genome size (pg) of Triturus arntzeni with that of other groups of Triturus newts. From left to right, the groups are T. pygmaeus (n = 1), T. marmoratus (n = 2), T. dobrogicus (n = 115), T. cristatus (n = 183), T. carnifex (n = 3), T. macedonicus (n = 6), T. arntzeni (white; n = 13), and western (n = 2), central (n = 2) and eastern (n = 58) T. karelinii s. l. nuclear gene pool (colors correspond to those in Fig. 1). Shown are median, quartiles and range. Data are taken from (Litvinchuk et al., 1999; Litvinchuk et al., 2007) and provided in online Appendix 3. Note that the genome size of T. arntzeni is dissimilar to T. karelinii s. l. and is not different from that of T. macedonicus and T. carnifex.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
1 (by plazi, 2016-04-14 14:15:03)
2 (by ImsDioSync, 2016-12-21 15:21:57)
3 (by ImsDioSync, 2016-12-21 15:22:31)
4 (by ExternalLinkService, 2019-09-26 15:08:25)
5 (by ExternalLinkService, 2021-11-09 12:08:37)
6 (by ExternalLinkService, 2021-11-09 15:17:53)
7 (by plazi, 2023-10-26 16:05:51)