Salvia miltiorrhiza, Bunge, Bunge
|
publication ID |
https://doi.org/10.1016/j.phytochem.2021.112932 |
|
DOI |
https://doi.org/10.5281/zenodo.8269776 |
|
persistent identifier |
https://treatment.plazi.org/id/03F987D2-FFF8-FFF3-AE7B-FE06FF01FE27 |
|
treatment provided by |
Felipe |
|
scientific name |
Salvia miltiorrhiza |
| status |
|
2.1. Identification of NAC family genes and phylogenetic analysis in S. miltiorrhiza View in CoL View at ENA
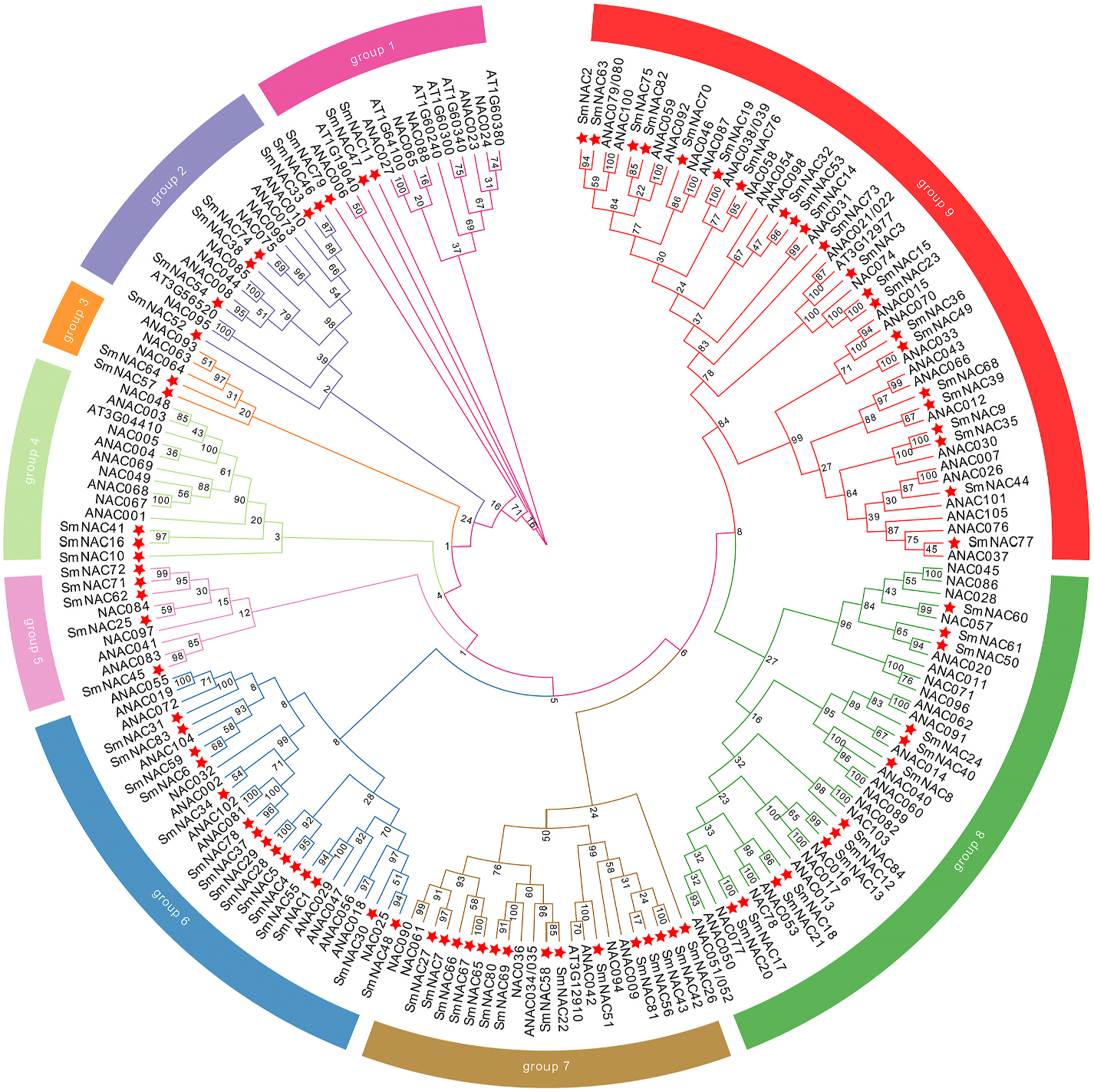
In this study, BLASTP and HMM searches using the NAC protein sequences of Arabidopsis as query were performed to broadly identify S. miltiorrhiza NAC family members. A total of 84 NAC proteins were identified in the S. miltiorrhiza genome. They were named Sm- NAC4 - Sm- NAC84 is based in accordance with gene IDs in the newly sequenced genome. Sm- NAC1 - Sm- NAC3 had been identified previously ( Zhang and Liang, 2019; Yin et al., 2020; Zhu et al., 2019b); therefore, their names were maintained (Supplementary Table S1). The coding DNA and protein sequences of Sm-NAC gene family members are given in Supplementary Files S1–S2. The protein sequence lengths of the 84 Sm-NACs are quite different (Supplementary Table S1). The longest is 794 bp (Sm-NAC79), and the shortest is only 120 bp (Sm-NAC27). The molecular weights range from 14.01 kDa (Sm-NAC27) to 89.74 kDa (Sm-NAC79), and the isoelectric points (pI) range from 4.53 (Sm-NAC83) to 9.73 (Sm-NAC4). The locations of the 84 Sm -NAC genes on the chromosomes of S. miltiorrhiza are shown in Supplementary Figure S1 View Fig , and all 84 Sm -NAC genes, except for Sm -NAC4 that is on the scaffold, are distributed among the eight chromosomes. There was no significant correlation between the number of Sm -NAC genes present per chromosome and the chromosomal length. To investigate the phylogenetic relationships among the 84 Sm-NACs, a phylogenetic tree was constructed by combining Sm-NACs with Arabidopsis NAC proteins (AtNACs). The 84 Sm-NACs were divided into nine families (Groups 1–9) ( Fig. 1 View Fig ). There was an unequal distribution of Sm-NACs among the groups, with the Group 9 subfamily, containing 22 genes, having the most members, followed by Groups 7 and 6, each containing 15 proteins. The Group 3 subfamily had the least members, containing only two genes.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |