Arcuseries minima, Kim & Chae & Min, 2019
|
publication ID |
https://doi.org/ 10.4467/16890027AP.19.005.10836 |
|
DOI |
https://doi.org/10.5281/zenodo.11094689 |
|
persistent identifier |
https://treatment.plazi.org/id/038D87C6-FFA2-226C-6F10-2BDF6FD8A2A0 |
|
treatment provided by |
Felipe |
|
scientific name |
Arcuseries minima |
| status |
sp. nov. |
Arcuseries minima sp. nov.
ZooBank: urn:lsid:zoobank.org:act:F17B7069-8B86-4A70-9654-42F85CDA0AC7
Diagnosis. Size in vivo 40–55 × 20–30 μm; elliptical in shape, highly flexible, and contractile; 20–27 macro-nuclear nodules and 1–3 micronuclei; contractile vacuole lacking. Three type of cortical granules: (1) colorless, spherical shape, scattered, ca. 3 μm; (2) yellowish, clustered around cirri and dorsal bristles, ca. 1 μm; (3) colorless, scattered, ca. 0.5 μm. About 14 adoral membranelles; 3 frontal, 1 buccal, 2 frontoterminal, 5–10 midventral, 2 pretransverse, and 5 or 6 transverse cirri; 1 left (9–13 cirri) and 1 right (9–14 cirri) marginal row; 3 dorsal kineties.
Type locality. Seawater (salinity, 33‰; water temperature, 27.1℃) taken from Jeju province, South GoogleMaps Korea (33°14’9.21”N; 126°35’55.38”E) in August 2017.
Type specimens. The slide ( NIBRPR0000109740 ) containing the holotype specimen and two slides ( NIBRPR0000109741 , NIBRPR0000109742 ) including protargol-stained specimens have been deposited in the National Institute of Biological Resources ( NIBR), Incheon, South Korea .
Etymology. The species-group name, minima , is the feminine version of the Latin adjective minimus (smallest) to indicate the small body size of the new species.
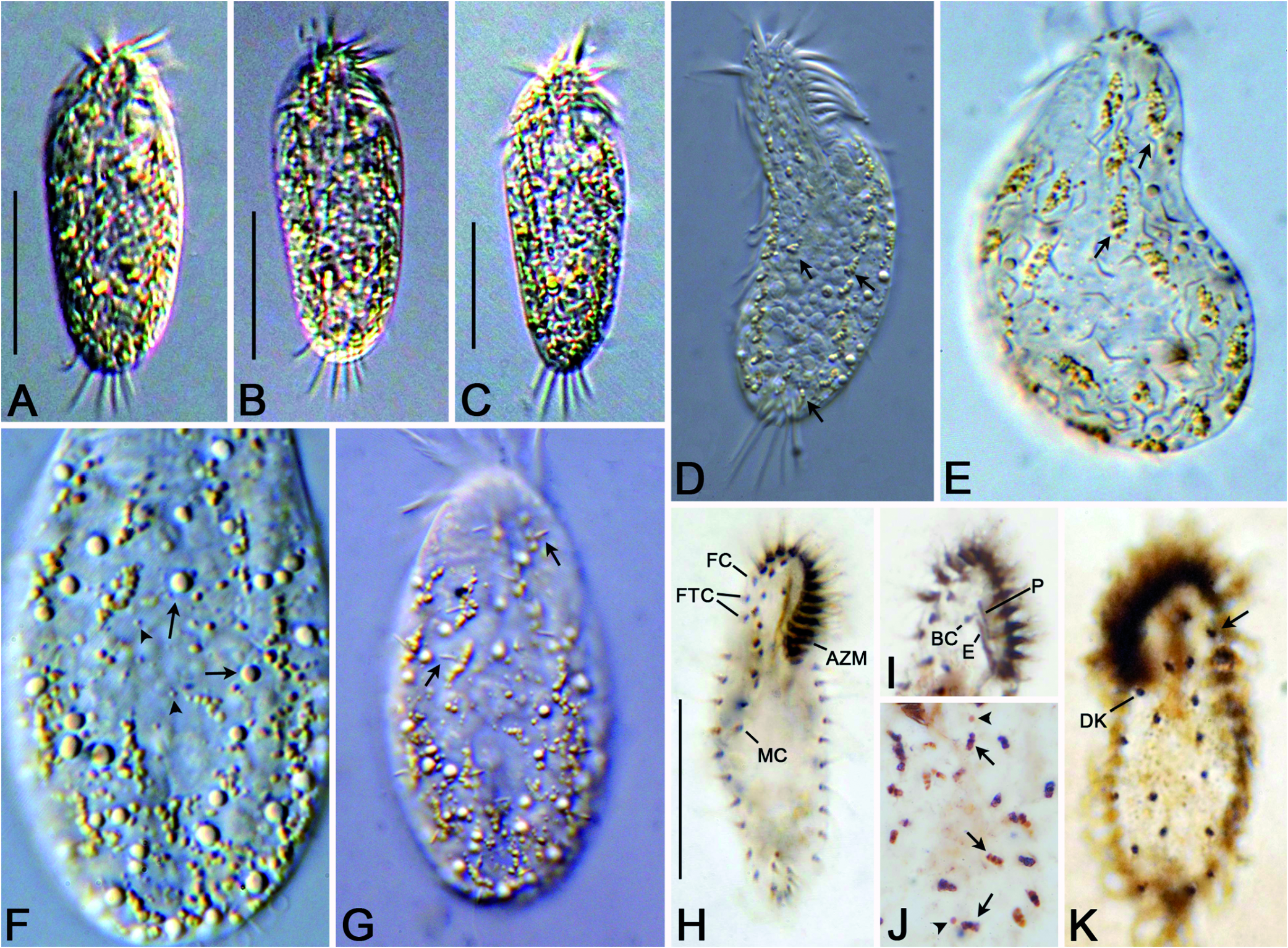
Description: Size in vivo 40–55 × 20–30 μm (n=15), about 34 × 11 μm in protargol preparations; body highly flexible, contractile, oval to elongated elliptical, left and right cell margins slightly convex, both ends rounded, and cell color yellowish to grayish at low magnification ( Figs. 1A, F View Fig , 2A–C, H View Fig ). Macronuclear nodules size about 3.3 × 1.9 μm in protargol impregnation, 20–27 in number; one to three micronuclei of size about 1.6 × 1.4 μm in protargol impregnation ( Figs. 1G View Fig , 2J View Fig ). Contractile vacuole lacking. Three types of cortical granules: type I (large-sized), colorless, spherical, about 3 μm in size, irregularly distributed on dorsal surfaces; type II (medium-sized), yellowish, spherical, about 1 μm in diameter, clustered around cirri and dorsal bristles; type III (small-sized), colorless, irregularly distributed on dorsal surfaces, less than 0.5 μm in diameter ( Figs. 1B–E View Fig , 2D–G View Fig ). Cytoplasm colorless, with 3–5 μm sized food vacuoles. Feeds on bacteria.
Adoral zone of membranelles occupies about 31.5% of cell length in protargol preparations, base of the largest membranelles is about 3.5 μm long, composed of 13–16 membranelles. Paroral and endoral membranes side by side, endoral membrane longer than paroral membrane ( Figs. 1F View Fig , 2H, I View Fig ). All cirri are relatively fine, generally 6–8 μm long in vivo except frontal and transverse cirri. Three slightly enlarged frontal cirri about 10 μm in length, the rightmost cirrus located behind distal end of the adoral zone of membranelles. Two frontoterminal cirri behind the rightmost frontal cirrus, buccal cirrus on the anterior of endoral membrane, and two pretransverse cirri located ahead the transverse cirri. Five or six slightly enlarged transverse cirri, 10–12 μm in length. Midventral complex consists of two to five midventral pairs (five to ten cirri), arranged in a zigzag pattern, commencing near buccal cirrus and terminating on or above midbody. Two marginal rows, one left (9–13 cirri) and one right (9–14 cirri) commencing behind the posterior end of the buccal field, and both rows non-confluent posteriorly ( Figs. 1A, F View Fig , 2H, I View Fig ).
Invariably three dorsal kineties, cilia about 3 μm long in vivo: kineties 1 and 2 composed of four to six bristles, anteriorly shortened; almost bipolar kinety 3 composed of five to seven bristles. One pair of basal bodies located ahead of the right marginal cirral row ( Figs. 1G View Fig , 2K View Fig ).
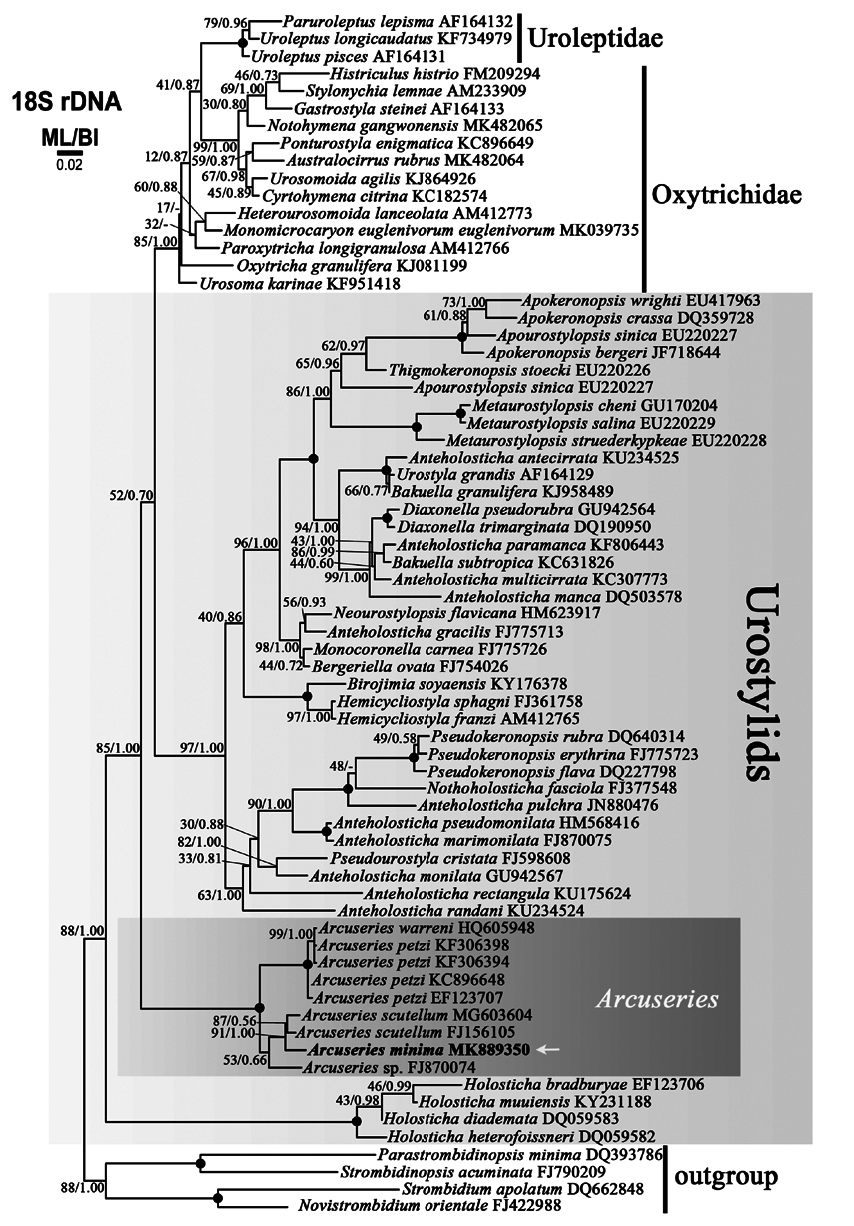
Molecular analyses: The 18S rDNA sequence of Arcuseries minima sp. nov. is 1,598 bp in length and has a GC content of 46.9%. The sequence of A. minima was deposited in GenBank with accession number MK 889350. BI and ML analyses produced similar topologies; thus, the ML tree was presented as our phylogenetic tree. In the gene tree, A. minima clusters with A. scutellum , and is a sister species to A. petzi and A. warreni ( Fig. 3 View Fig ). In addition, A. minima shows a shorter pairwise distance with A. scutellum (0.009, 14 or 15 differences in 1,598 nt) than with A. sp. (0.018, 29 differences in 1,598 nt), A. petzi (0.024 –0.027, 38–43 differences in 1,600 nt), and A. warreni (0.026, 42 differences in 1,600 nt) (Table 3).
| NIBR |
National Institute of Biological Resources |
| GC |
Goucher College |
| MK |
National Museum of Kenya |
| ML |
Musee de Lectoure |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |