Cephalotaxus harringtonia subsp. crude
|
publication ID |
https://doi.org/10.1016/j.phytochem.2014.09.021 |
|
DOI |
https://doi.org/10.5281/zenodo.10522582 |
|
persistent identifier |
https://treatment.plazi.org/id/0388C53D-FF94-2E76-FFC9-FF276A5FFE8F |
|
treatment provided by |
Felipe |
|
scientific name |
Cephalotaxus harringtonia subsp. crude |
| status |
|
2.1. Metabolomics analysis of the C. harringtonia crude extract transformed by the endophyte P. variabile
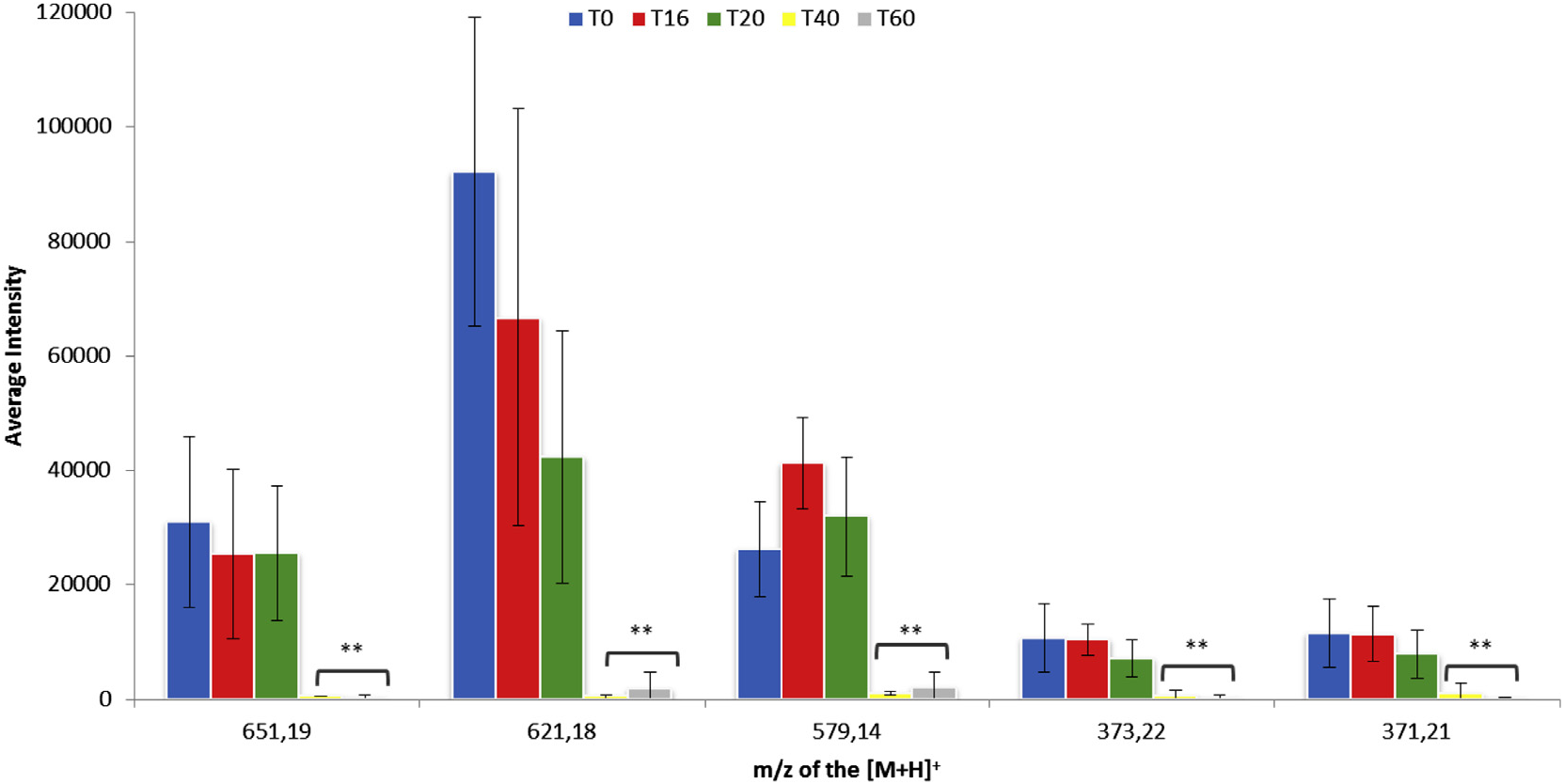
Biotransformations of the crude extract of C. harringtonia by the endophyte P. variabile were monitored by standardized LC– MS fingerprints at 16, 20, 40, and 60 h post inoculation (hpi). Using the XCMS software, the protonated ions of the biotransformed metabolites at the different hpi were compared to the control crude extract sample ( t = 0 h).
From these analyses more than one hundred protonated ions showed a decreasing intensity in the course of the biotransformation. To analyze the most abundant and relevant ions, we selected only those with an intensity superior to 10,000 and a relative fold change, as defined by XCMS, superior to 100. With these criteria, five metabolites appeared to be transformed predominantly by the fungus. Indeed, the pseudomolecular ions [ M +H] + at m/z 371.21, 373.22, 579.14, 621.18 and 651.19 underwent a significant decrease of intensity 40 h after incubation with the mycelium of P. variabile ( Fig. 1 View Fig ). Among these metabolites, those at m/z 579.14, 621.18 and 651.19 were the most transformed.
2.2. Isolation and characterization of the main compounds metabolized by P. variabile
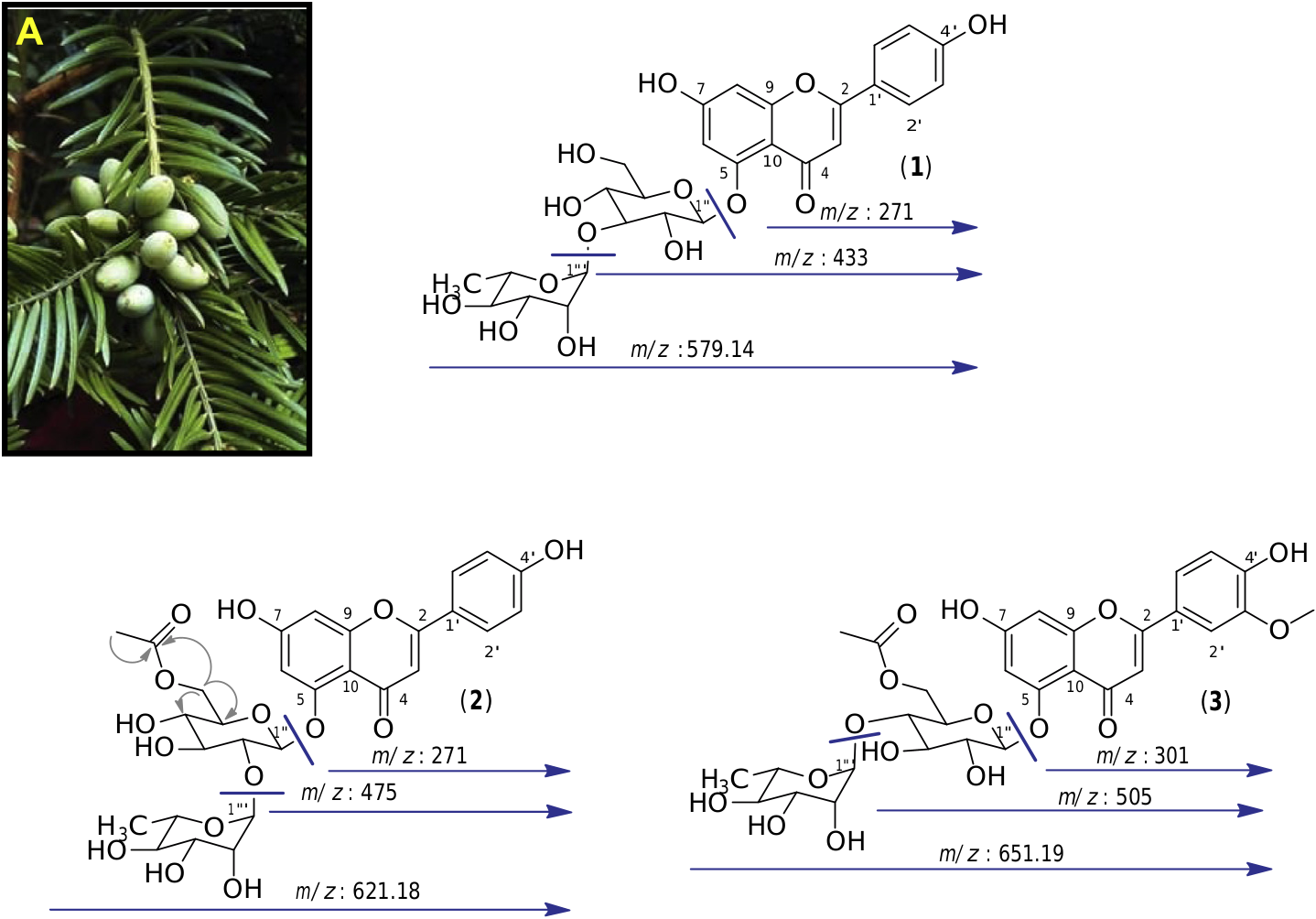
We started by analyzing the compounds that had been most transformed generating ions at m/z 579.14, 621.18 and 651.19. We isolated them from the crude extract by semi-preparative HPLC guided by MS detection, giving the tree main metabolites at m/z 579.14, 621.18 and 651.19. The analysis of 1D and 2D NMR spectra, along with the MS / MS fragmentations and bibliographic data, allowed unambiguous characterization of these compounds. The collision-induced dissociation (CID) experiments on compound ( 1), which displays a pseudomolecular protonated ion [ M +H] + at m/z: 579.14, led to two main fragments at m/z 433 and 271, corresponding to the successive loss of 146 u and 162 u. These were attributed to the loss of a deoxyhexose and a hexose suggesting that compound 1 is a diglycoside ( Fig. 2 View Fig ).
The 1 H MNR spectrum of compound 1 in CD 3 OD displayed an olefinic signal at dH 6.51 (1H, s, H-3) characteristic of a flavone structure. This compound appeared to be dihydroxylated at position 4 0 and 7, as indicated by signals at dH 7.81 (2H, d, J = 8.7 Hz, H-2 0, H-6 0), dH 6.92 (2H, d, J = 8.7 Hz, H-3 0, H-5 0) which are characteristic of a p -hydroxyphenyl ring, and at dH 6.63 (1H, d, J = 2.2 Hz, H-8) and dH 6.60 (1H, d, J = 2.2 Hz, H-6) which reveal the presence of a phloroglucinol ring. Two anomeric protons at dH 5.37 (1H, d, J = 1.5 Hz, H-1 000) and dH 5.30 were also (1H, d, J = 6.6 Hz, H-1 00) present, characteristic of the diglycoside part, and a methyl proton at dH 1.12 (3H, d, J = 7.0 Hz, H-6 000) showing the deoxyhexose nature of one of the sugar unit. The interglycosidic linkage as well as the attachment position of the diglycoside to the aglycone was confirmed by rigorous analysis of the 1 H A 1 H COSY and 1 H A 13 C HMBC correlations, both showing 2-bond and 3-bond scalar couplings. The configuration of the glycosidic linkage of the glucopyranoside and rhamnopyranoside were determined to be β and oi, respectively, on the basis of the 3 J 1 00 –2 00 (6.6 Hz) and 3 J 1 000 –2 000 (1.5 Hz) values of the anomeric protons. Thus, from these results and from comparison with bibliographic data, the structure of this compound was assigned as apigenin 5- O -a- L- rhamnopyranosyl-(1 + 3)- β -D- glucopyranoside ( 1) which was previously isolated from a Cephalotaxus tree ( Bae et al., 2007) ( Fig. 2 View Fig ).
In the same way, we identified by one-and two-dimensional NMR as well as MS / MS experiments, compound 2 which displayed a pseudomolecular protonated ion [ M +H] + at m/z: 621.18 as apigenin 5- O -a- L- rhamnopyranosyl-(1 + 2)-(6 00 - O -acetyl)- β -D- glucopyranoside, also known as euryanoside ( 2) ( Inada et al., 1989). Compound 3 with an m/z 651.19 was assigned as chrysoeriol 5- O - a- L- rhamnopyranosyl-(1 + 4)-(6 00 - O -acetyl)- β -D- glucopyranoside, another flavone glycoside previously isolated from a Cephalotaxus tree ( Zhang et al., 2011) ( Fig. 2 View Fig ).
2.3. Preparative scale biotransformation of the isolated compounds 1– 3
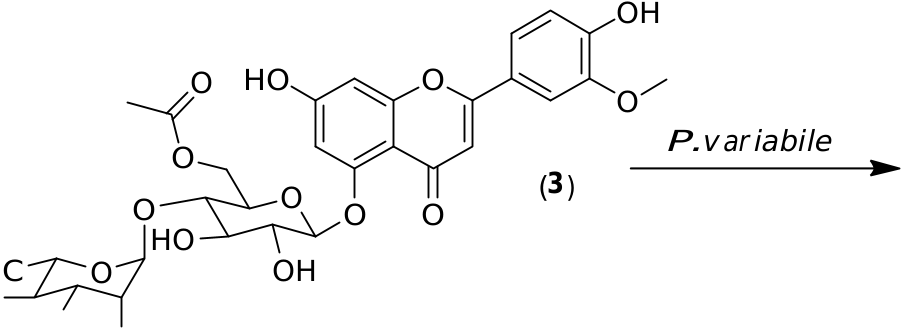
Preparative scale biotransformations of the pure glycosylated flavonoids 1–3 obtained from the crude extract of C. harringtonia were performed. Compounds 1–3 were added to P. variabile in a phosphate buffer. In all cases, large decreases of the substrate concentrations were observed by HPLC monitoring after 24 h of inoculation. The resulting products were isolated by chromatography before NMR and MS analyses. From these, we saw deglycosylation reactions in all cases, leading to two different aglycones. Indeed, from flavonoids 1 and 2, apigenin ( 4) was characterized while, from flavonoid 3, the small amount of product ( 5) could be assigned as chrysoeriol by comparison with a commercial standard ( Fig. 3 View Fig ).
2.4. Flavones 4 and 5 increase the growth of P. variabile hyphae
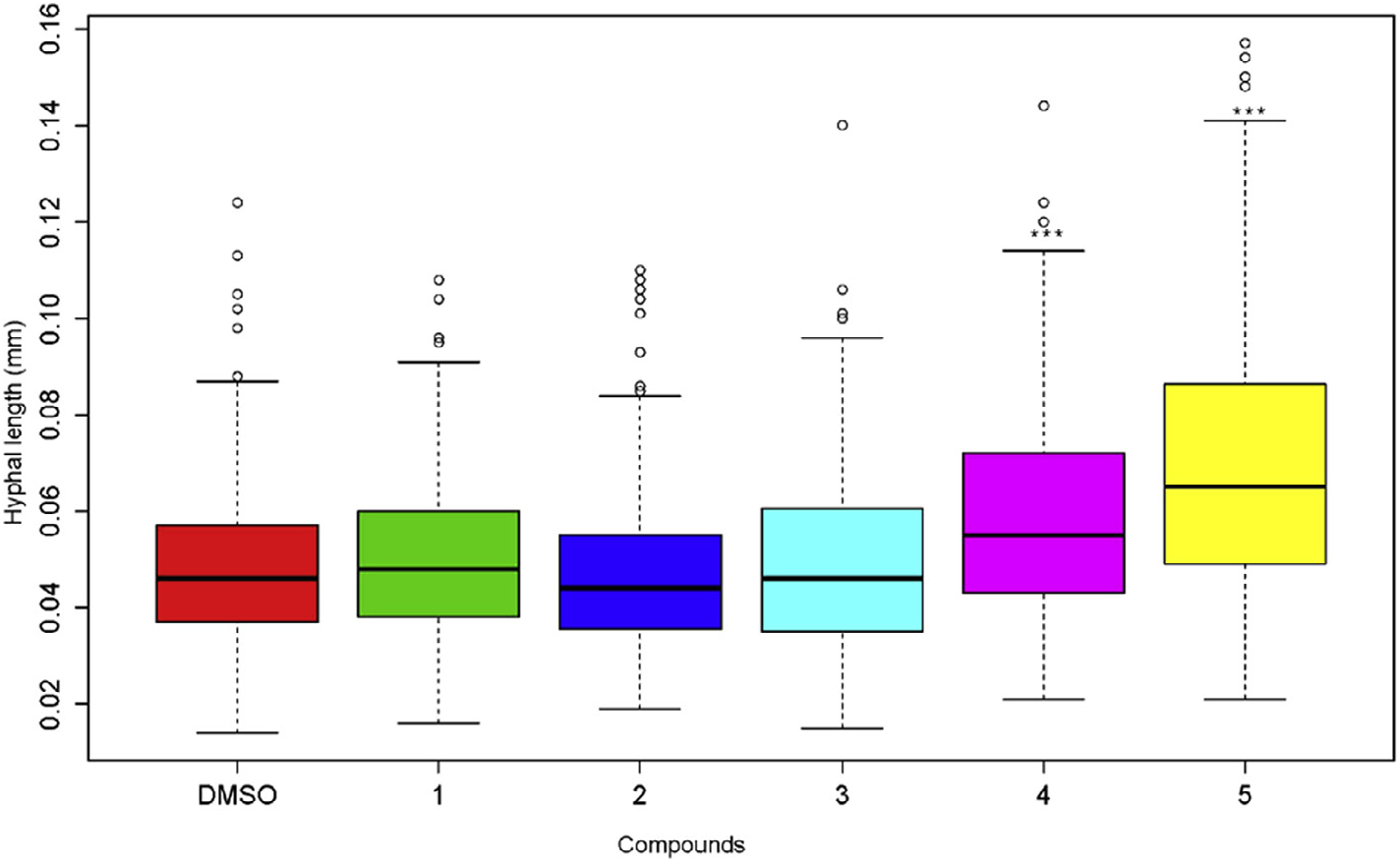
We investigated the respective impact of glycosylated flavonoids ( 1–3) and aglycones ( 4, 5) on the germination and hyphal growth of the spores of P. variabile . While no effect was observed on spore germination in any case (data not shown), we clearly observed ( Fig. 4 View Fig ) that after 24 h, the two aglycones apigenin ( 4) and, mostly, chrysoeriol ( 5) caused a significant improvement of the hyphal growth compared to the corresponding glycosylated compounds ( 1–3). No inhibition of spore germination and hyphal growth of the spore of P. variabile was observed with either the glycosylated flavonoids, or the corresponding flavonoids.
| MS |
Herbarium Messanaensis, Università di Messina |
| M |
Botanische Staatssammlung München |
| A |
Harvard University - Arnold Arboretum |
| C |
University of Copenhagen |
| O |
Botanical Museum - University of Oslo |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |