Rasopone
|
publication ID |
https://doi.org/10.1093/isd/ixaa004 |
|
DOI |
https://doi.org/10.5281/zenodo.3847174 |
|
persistent identifier |
https://treatment.plazi.org/id/9C548790-FFEC-FFAE-FF69-414EFBD2FF29 |
|
treatment provided by |
Felipe (2020-05-14 13:22:24, last updated 2024-11-26 03:50:06) |
|
scientific name |
Rasopone |
| status |
|
Rasopone JTL034
( Fig. 7 View Fig ; Supp Fig. S37 [online only])
Geographic range. Mexico (Puebla, Veracruz), Honduras.
Diagnosis
Mandible smooth and shining or very faintly striate; anterior clypeal margin truncate; side of head with short erect setae; petiolar node moderately tapering, intermediate between cuboidal and scale-like. Three species are within geographic and size range of R. JTL034:
Rasopone guatemalensis ( Fig. 8 View Fig ; Supp Figs. S14 and S15 [online only]): no known local sympatry, but ranges overlap; petiolar node slightly more tapering, scale-like; scape longer (mean SI 86 vs 76); head narrower (mean CI 86 vs 93).
Rasopone politognatha ( Fig. 9 View Fig ; Supp Figs. S28 and S29 [online only]): no known local sympatry, but ranges overlap; side of head bare; scape slightly longer (mean SI 82 vs 76).
Rasopone JTL035 ( Fig. 9 View Fig ; Supp Fig. S38 [online only]): no known local sympatry but in close proximity in Sierra de Los Tuxtlas, possibly segregating by elevation; somewhat larger and darker red brown; side of head bare.
Measurements, worker: HW 1.11 (1.04–1.15, 3); HL 1.19 (1.12– 1.23, 3); SL 0.85 (0.79–0.91, 3); PTH 0.73 (0.68–0.77, 3); PTL 0.42 (0.39–0.45, 3); CI 93 (92–93, 3); SI 77 (75–79, 3); PTI 58 (56–61, 3).
Biology
This species occurs in wet to moist forest, with records from 170 to 980 m elevation. It is known from four widely separated localities. The three known worker specimens are from Winkler samples of forest floor litter and rotten wood. A male was collected 14 June 2010 in a Malaise trap.
Comments
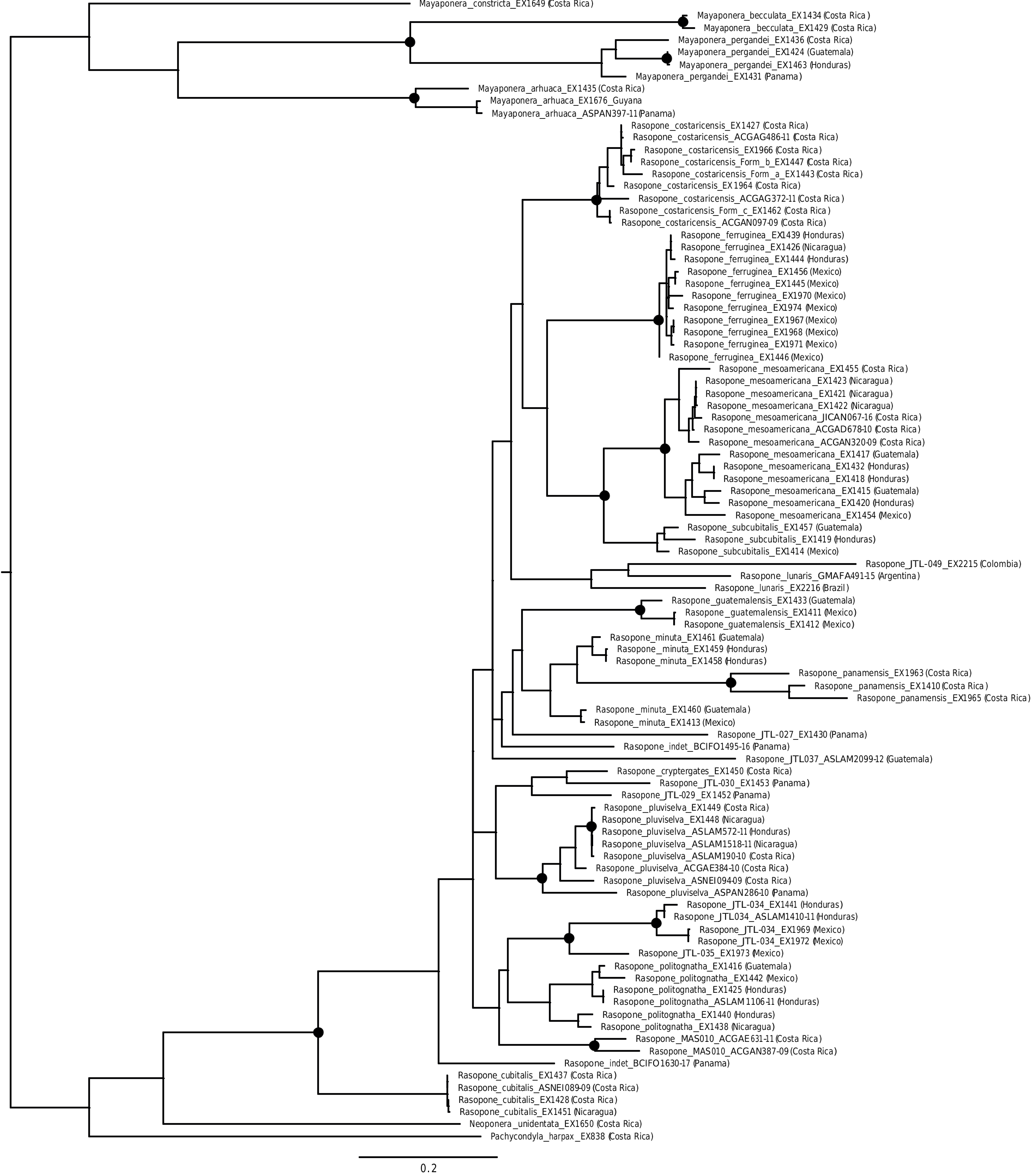
UCE results unite three workers from widely separated localities. The sister taxon is the single specimen of R. JTL035, which appears to be a more montane version of R. JTL034 that is slightly larger and darker. The specimen of R. JTL035 is from a cloud forest site in Sierra de Los Tuxtlas, while one of the specimens of R. JTL034 is from just a few kilometers away, in the lowland rainforest of Los Tuxtlas Biological Station. However, the Los Tuxtlas specimen of R. JTL034 forms a clade with specimens from Puebla ( Mexico) and Honduras. COI data unite a male from Tela, Honduras, with the three worker specimens ( Fig. 3 View Fig ).
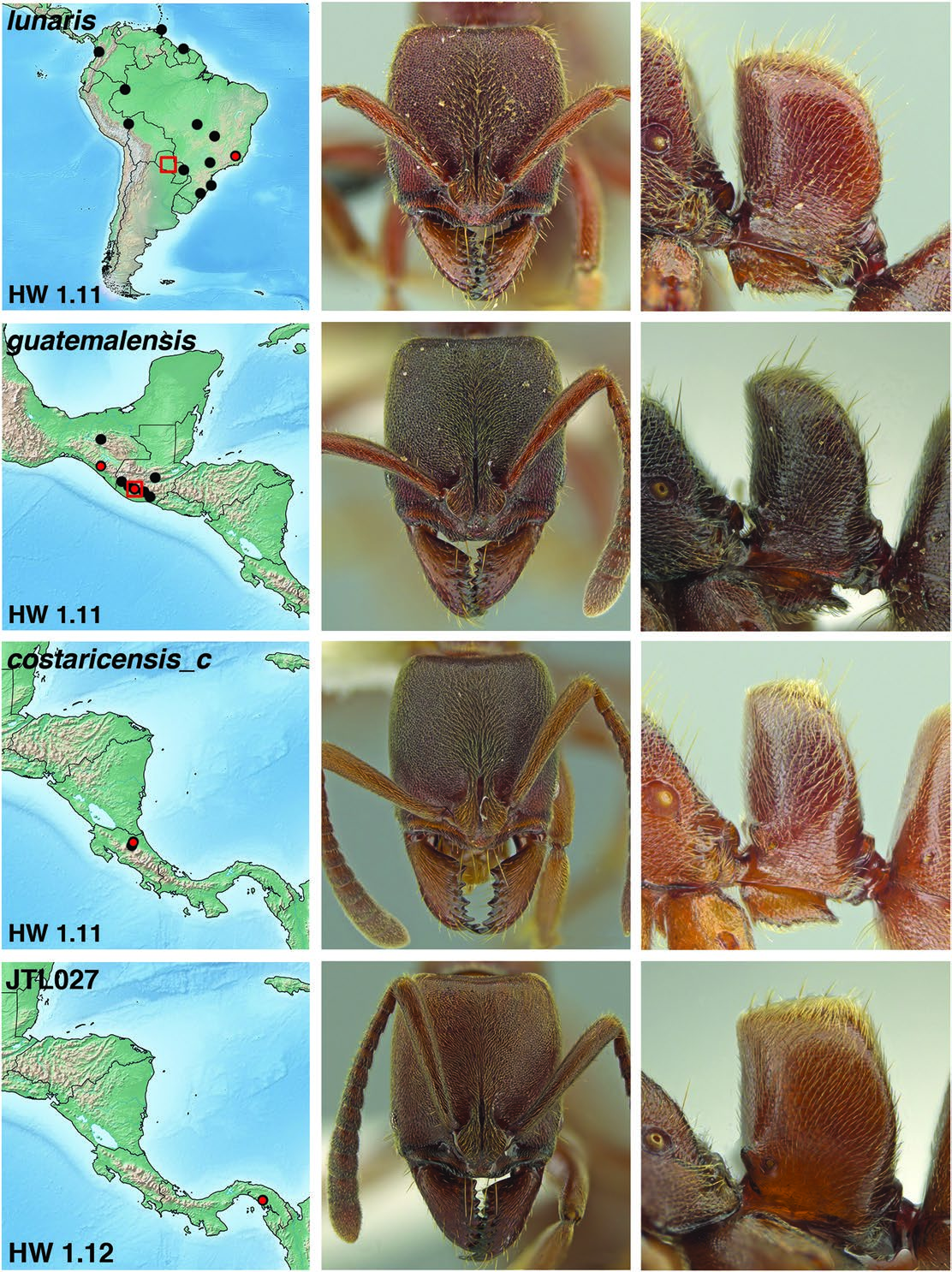
Fig. 7. Distribution map, face view, and lateral view of petiole of Rasopone MAS010 (worker, Costa Rica, 08COSTA-1723), R. cryptergates (worker, Costa Rica, INB0003660648), R. JTL042 (worker, Colombia, MCZ-ENT00716611),and R. JTL034 (worker, Mexico, CASENT0640282). On distribution maps, red dots are sites with UCE sequence data. Red boxes are type locality.
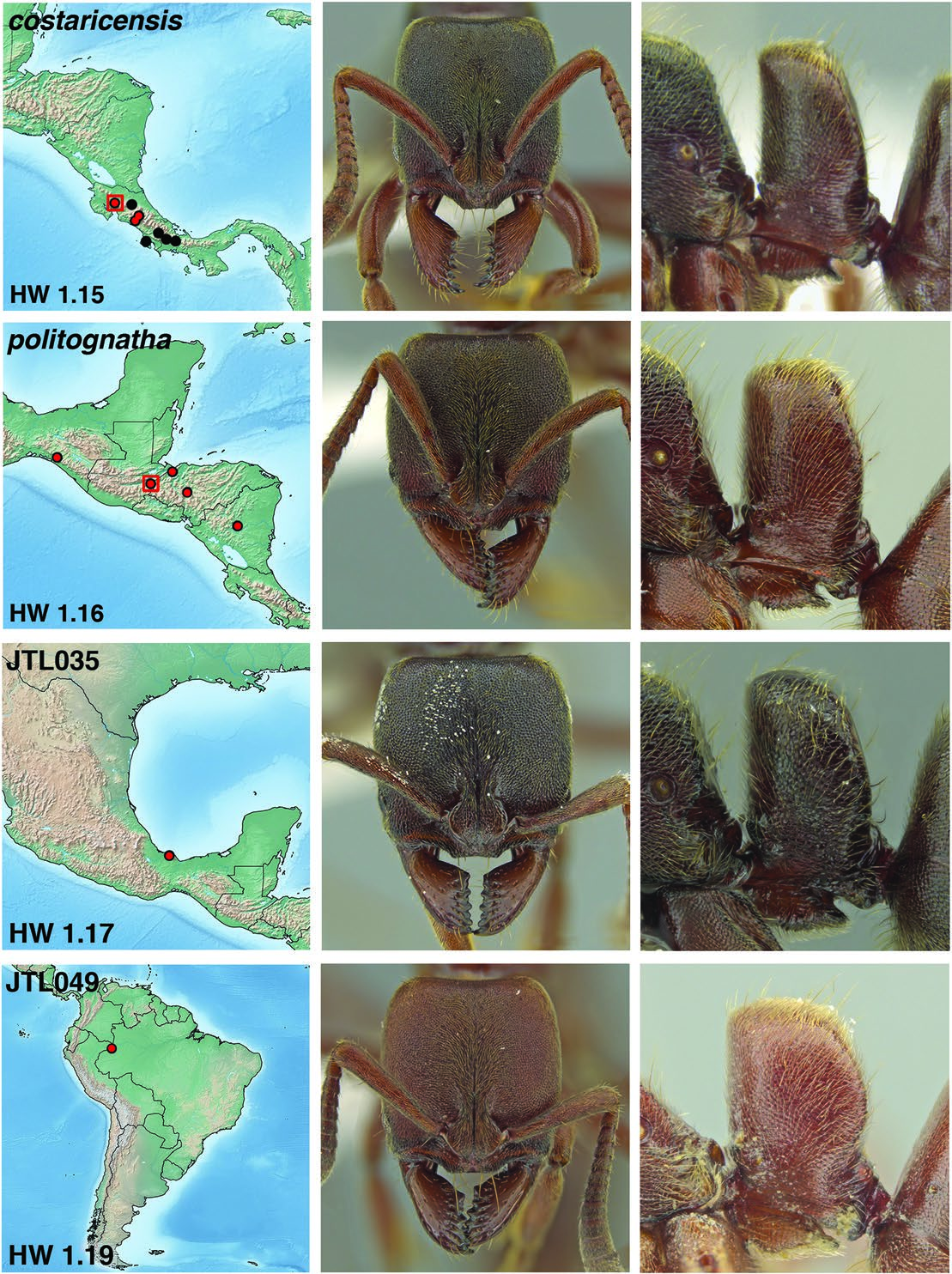
Fig. 8. Distribution map, face view, and lateral view of petiole of Rasopone lunaris (worker, Brazil, Minas Gerais CASENT0644556), R. guatemalensis (holotype worker),R. costaricensis form c (worker,Costa Rica,INB0003659307),and R. JTL027 (worker,Panama,CASENT0633216).On distribution maps,red dots are sites with UCE sequence data. Red boxes are type locality.
Fig. 9. Distribution map, face view, and lateral view of petiole of Rasopone costaricensis (holotype worker), R. politognatha (holotype worker), R. JTL035 (worker, Mexico, CASENT0640453), and Rasopone JTL049 (worker, Colombia, CASENT0644557). On distribution maps, red dots are sites with UCE sequence data. Red boxes are type locality.
Fig. 3. Phylogenetic relationships among a curated set of COI barcode sequences for Rasopone. Black samples were sequenced for UCEs. Red samples were downloaded from the BOLD database.The tree was inferred using IQ-TREE with the data partitioned by codon position. Black circles on nodes indicate high support, which we define as ≥95% ultrafast bootstrap support and ≥95% SH-like branch support.Terminal names match taxonomic changes proposed in paper and provide useful sample identifiers (e.g., extraction codes [EX#] or BOLD process IDs).A complete,unpruned COI tree is available in Supp Fig. S1 (online only).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
1 (by felipe, 2020-05-14 13:22:24)
2 (by ExternalLinkService, 2020-05-14 13:33:21)
3 (by ExternalLinkService, 2020-05-26 00:19:55)
4 (by ExternalLinkService, 2021-10-20 04:05:05)
5 (by ExternalLinkService, 2021-10-20 08:51:27)
6 (by plazi, 2023-10-31 14:21:32)